유용한 함수들
select(), mutate(), filter(), rename() : 기본 tidyverse verbs
rowSums(), rowMeans() : composite 변수들의 합 또는 평균을 구함
factor() : 카테고리 변수의 변환
앞서 다운받은 데이터: altruism.csv 파일 링크
library(tidyverse)
# import data
helping <- read_csv("data/altruism.csv")
helping |> print()
# A tibble: 120 x 12
id pho_1 pho_2 pho_3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 80 80 70
2 2 58 62 NA 0 2003 62 58 59 57 56
3 3 100 50 50 NA 2003 90 51 51 51 52
4 4 77 77 64 1 2004 66 72 88 82 67
5 5 NA NA NA NA NA NA NA NA NA NA
6 6 100 75 100 0 2004 100 60 70 55 70
# i 114 more rows
# i 1 more variable: emp_q26 <dbl>
변수명 수정
rename()
helping |>
rename(ph1 = pho_1, ph2 = pho_2, ph3 = pho_3) |>
print()
# A tibble: 120 x 12
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 80 80 70
2 2 58 62 NA 0 2003 62 58 59 57 56
3 3 100 50 50 NA 2003 90 51 51 51 52
4 4 77 77 64 1 2004 66 72 88 82 67
5 5 NA NA NA NA NA NA NA NA NA NA
6 6 100 75 100 0 2004 100 60 70 55 70
# i 114 more rows
# i 1 more variable: emp_q26 <dbl>
변형 후에는 꼭 변수에 assign!
helping <- # 원래 데이터에 overwrite
helping |>
rename(ph1 = pho_1, ph2 = pho_2, ph3 = pho_3)
helping |> print()
# A tibble: 120 x 12
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 80 80 70
2 2 58 62 NA 0 2003 62 58 59 57 56
3 3 100 50 50 NA 2003 90 51 51 51 52
4 4 77 77 64 1 2004 66 72 88 82 67
5 5 NA NA NA NA NA NA NA NA NA NA
6 6 100 75 100 0 2004 100 60 70 55 70
# i 114 more rows
# i 1 more variable: emp_q26 <dbl>
문항을 더할 때
rowSums(row, na.rm = TRUE) 함수를 이용하는 것이 직접 덧셈보다 더 적절함
ph1, ph2, ph3 세 문항을 더하려면,
# 먼저 문항을 선택/확인
helping |>
select(ph1:ph3) |> # position!
print()
# A tibble: 120 x 3
ph1 ph2 ph3
<dbl> <dbl> <dbl>
1 95 95 95
2 58 62 NA
3 100 50 50
4 77 77 64
5 NA NA NA
6 100 75 100
# i 114 more rows
helping |>
select(ph1:ph3) |>
rowSums(na.rm = TRUE) |>
print()
[1] 285 120 200 218 0 275 257 178 256 189 215 226 209 246 159 197 205 225
[19] 150 195 44 0 0 125 225 211 270 176 241 205 0 220 98 79 143 165
[37] 49 294 300 292 101 285 208 230 255 150 299 188 208 205 138 267 187 300
[55] 195 300 236 59 226 193 213 250 32 228 250 300 300 190 230 281 196 268
[73] 240 250 39 233 211 198 199 234 300 215 240 9 261 209 281 201 270 255
[91] 177 235 161 0 242 151 182 170 3 222 172 194 300 300 293 238 243 260
[109] 197 294 280 195 255 1 162 278 176 262 300 164
helping["phone"] <- # "phone"이라는 새로운 변수에 assign!
helping |>
select(ph1:ph3) |>
rowSums(na.rm = TRUE)
helping |> print(width = Inf)
# A tibble: 120 x 13
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 80 80 70
2 2 58 62 NA 0 2003 62 58 59 57 56
3 3 100 50 50 NA 2003 90 51 51 51 52
4 4 77 77 64 1 2004 66 72 88 82 67
5 5 NA NA NA NA NA NA NA NA NA NA
6 6 100 75 100 0 2004 100 60 70 55 70
emp_q26 phone
<dbl> <dbl>
1 70 285
2 59 120
3 100 200
4 69 218
5 NA 0
6 90 275
# i 114 more rows
다음과 같이 직접 더하는 것은 부적절
helping |>
mutate(phone = ph1 + ph2 + ph3)
# id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
# <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
# 1 1 95 95 95 1 2004 80 NA 80 80 70
# 2 2 58 62 NA 0 2003 62 58 59 57 56
# 3 3 100 50 50 NA 2003 90 51 51 51 52
# 4 4 77 77 64 1 2004 66 72 88 82 67
# 5 5 NA NA NA NA NA NA NA NA NA NA
# 6 6 100 75 100 0 2004 100 60 70 55 70
# emp_q26 phone
# <dbl> <dbl>
# 1 70 285
# 2 59 NA
# 3 100 200
# 4 69 218
# 5 NA NA
# 6 90 275
# # … with 114 more rows
문항을 평균낼 때
rowMeans(row, na.rm = TRUE) 함수를 이용하는 것이 적절함
# 먼저, 평균을 낼 문항을 선택/확인
helping |>
select(emp_q20, emp_q22:emp_q26) |> # ":" operator와 "," 섞어써도 무방
print()
# A tibble: 120 x 6
emp_q20 emp_q22 emp_q23 emp_q24 emp_q25 emp_q26
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 80 NA 80 80 70 70
2 62 58 59 57 56 59
3 90 51 51 51 52 100
4 66 72 88 82 67 69
5 NA NA NA NA NA NA
6 100 60 70 55 70 90
# i 114 more rows
helping["persp"] <- helping |> # "persp"라는 새로운 변수에 assign!
select(emp_q20, emp_q22:emp_q26) |>
rowMeans(na.rm = TRUE)
helping |> print(width = Inf)
# A tibble: 120 x 14
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 80 80 70
2 2 58 62 NA 0 2003 62 58 59 57 56
3 3 100 50 50 NA 2003 90 51 51 51 52
4 4 77 77 64 1 2004 66 72 88 82 67
5 5 NA NA NA NA NA NA NA NA NA NA
6 6 100 75 100 0 2004 100 60 70 55 70
emp_q26 phone persp
<dbl> <dbl> <dbl>
1 70 285 76
2 59 120 58.5
3 100 200 65.8
4 69 218 74
5 NA 0 NaN
6 90 275 74.2
# i 114 more rows
표준화 및 중심화
- 표준화(standardize):
scale(x) 함수를 이용
- 중심화(center):
scale(x, scale = FALSE) 함수를 이용
helping |>
mutate(
phone_z = scale(phone) |> as.vector(), # scale()은 matrix로 반환; vector 변환 필요
persp_z = scale(persp) |> as.vector()
) |>
print(n = 3, width = Inf)
# A tibble: 120 x 16
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 80 80 70
2 2 58 62 NA 0 2003 62 58 59 57 56
3 3 100 50 50 NA 2003 90 51 51 51 52
emp_q26 phone persp phone_z persp_z
<dbl> <dbl> <dbl> <dbl> <dbl>
1 70 285 76 1.05 0.104
2 59 120 58.5 -1.00 -0.981
3 100 200 65.8 -0.00643 -0.527
# i 117 more rows
across() 함수를 이용하면 여러 변수를 한 번에 변환할 수 있음
helping |>
mutate(across(
.cols = c(phone, persp),
.fns = ~ (scale(.) |> as.vector()),
.names = "{.col}_z"
)) |> # 변수명을 일괄 변경: 변수명 + "_z"
print(n = 3, width = Inf)
# A tibble: 120 x 16
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 80 80 70
2 2 58 62 NA 0 2003 62 58 59 57 56
3 3 100 50 50 NA 2003 90 51 51 51 52
emp_q26 phone persp phone_z persp_z
<dbl> <dbl> <dbl> <dbl> <dbl>
1 70 285 76 1.05 0.104
2 59 120 58.5 -1.00 -0.981
3 100 200 65.8 -0.00643 -0.527
# i 117 more rows
카테고리 변수 및 연산
카테고리 변수는 R의 factor 타입으로 바꾸어 분석하는 것이 유리함.
간단한 연산은 직접 계산.
helping |>
mutate(
sex = factor(sex, levels = c(0, 1), labels = c("male", "female")), # factor 타입의 변수로 변환
age = 2023 - age # 출생년도로부터 나이 계산
) |>
print(width = Inf)
# A tibble: 120 x 14
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 female 19 80 NA 80 80 70
2 2 58 62 NA male 20 62 58 59 57 56
3 3 100 50 50 NA 20 90 51 51 51 52
4 4 77 77 64 female 19 66 72 88 82 67
5 5 NA NA NA NA NA NA NA NA NA NA
6 6 100 75 100 male 19 100 60 70 55 70
emp_q26 phone persp
<dbl> <dbl> <dbl>
1 70 285 76
2 59 120 58.5
3 100 200 65.8
4 69 218 74
5 NA 0 NaN
6 90 275 74.2
# i 114 more rows
행의 삭제
filter()를 활용
예를 들어, 5번째 행을 지우려면
helping |>
filter(!id == 5) |> # !는 not의 의미
print()
# 다시 helping에 assign 해야 수정됨!
# A tibble: 119 x 14
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 80 80 70
2 2 58 62 NA 0 2003 62 58 59 57 56
3 3 100 50 50 NA 2003 90 51 51 51 52
4 4 77 77 64 1 2004 66 72 88 82 67
5 6 100 75 100 0 2004 100 60 70 55 70
6 7 77 94 86 1 2004 91 93 85 91 73
# i 113 more rows
# i 3 more variables: emp_q26 <dbl>, phone <dbl>, persp <dbl>
여러 행을 지우려면?
%in% 응용
helping |>
filter(!id %in% c(1, 3, 5)) |> # !는 not의 의미
print()
# A tibble: 117 x 14
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2 58 62 NA 0 2003 62 58 59 57 56
2 4 77 77 64 1 2004 66 72 88 82 67
3 6 100 75 100 0 2004 100 60 70 55 70
4 7 77 94 86 1 2004 91 93 85 91 73
5 8 90 68 20 0 2004 67 66 31 67 63
6 9 100 79 77 0 2003 61 51 30 51 51
# i 111 more rows
# i 3 more variables: emp_q26 <dbl>, phone <dbl>, persp <dbl>
열의 삭제
select() 활용
emp_q23, emp_q25 두 열을 삭제
helping |>
select(-emp_q23, -emp_q25) |>
print()
# A tibble: 120 x 12
id ph1 ph2 ph3 sex age emp_q20 emp_q22 emp_q24 emp_q26 phone
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 80 70 285
2 2 58 62 NA 0 2003 62 58 57 59 120
3 3 100 50 50 NA 2003 90 51 51 100 200
4 4 77 77 64 1 2004 66 72 82 69 218
5 5 NA NA NA NA NA NA NA NA NA 0
6 6 100 75 100 0 2004 100 60 55 90 275
# i 114 more rows
# i 1 more variable: persp <dbl>
emp_q23부터 emp_q26 열을 삭제 (위치의 의미로)
helping |>
select(-(emp_q23:emp_q26)) |> # () 꼭 필요
print()
# A tibble: 120 x 10
id ph1 ph2 ph3 sex age emp_q20 emp_q22 phone persp
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 95 95 95 1 2004 80 NA 285 76
2 2 58 62 NA 0 2003 62 58 120 58.5
3 3 100 50 50 NA 2003 90 51 200 65.8
4 4 77 77 64 1 2004 66 72 218 74
5 5 NA NA NA NA NA NA NA 0 NaN
6 6 100 75 100 0 2004 100 60 275 74.2
# i 114 more rows
이상치 발견
Outliers을 찾는 방법은 다양하고 복잡한 테크닉을 요하기도 하는데, 앞으로 점차 익히게 될 것임
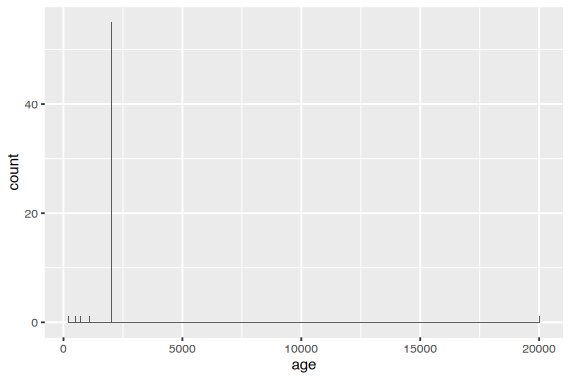
예를 들어, age에 잘못 기입한 경우가 있는데
helping <- read_csv("data/altruism.csv")
helping |>
ggplot(aes(x = age)) +
geom_histogram(binwidth = 100)
age는 출생년도를 물어봤으나 다른 답을 한 경우들이 있음
값은 2002 ~ 2004 사이가 정상이므로 filter()를 써서 확인해 볼 수 있음
helping |>
filter(age < 2002 | age > 2004) |>
print()
# A tibble: 7 x 12
id pho_1 pho_2 pho_3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 11 65 94 56 1 203 62 86 58 47 62
2 21 17 10 17 1 20004 0 6 1 0 4
3 43 90 88 30 1 507 100 78 62 100 78
4 52 100 82 85 1 723 87 83 89 100 88
5 59 76 86 64 0 709 100 93 67 94 79
6 108 75 100 85 1 2005 100 100 100 100 97
7 118 92 76 94 0 1108 55 51 51 60 53
# i 1 more variable: emp_q26 <dbl>
값을 수정
# 이상치에 대한 id를 우선 추출
ids_anomaly <- helping |>
filter(age < 2002 | age > 2004) |>
pull(id) # vector로 반환
ids_anomaly |> print()
[1] 11 21 43 52 59 108 118
# 모두 NA로 변경하는 경우
helping |>
# ifelse(조건, 참일 때 값, 거짓일 때 값)
mutate(
age = ifelse(age > 2004, 2004, age),
age = ifelse(age < 2002, NA, age)
# 대신, age = ifelse(age > 2004, 2004, ifelse(age < 2002, NA, age))
) |>
filter(id %in% ids_anomaly) |>
print()
# A tibble: 7 x 12
id pho_1 pho_2 pho_3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 11 65 94 56 1 NA 62 86 58 47 62
2 21 17 10 17 1 2004 0 6 1 0 4
3 43 90 88 30 1 NA 100 78 62 100 78
4 52 100 82 85 1 NA 87 83 89 100 88
5 59 76 86 64 0 NA 100 93 67 94 79
6 108 75 100 85 1 2004 100 100 100 100 97
7 118 92 76 94 0 NA 55 51 51 60 53
# i 1 more variable: emp_q26 <dbl>
# 직접 입력하는 방식
helping[helping$id %in% ids_anomaly, "age"] |> print()
# A tibble: 7 x 1
age
<dbl>
1 203
2 20004
3 507
4 723
5 709
6 2005
7 1108
# 직접 입력하는 방식
helping[helping$id %in% ids_anomaly, "age"] <- c(NA, 2004, NA, NA, NA, 2005, NA)
helping |>
filter(id %in% ids_anomaly) |>
print()
# A tibble: 7 x 12
id pho_1 pho_2 pho_3 sex age emp_q20 emp_q22 emp_q23 emp_q24 emp_q25
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 11 65 94 56 1 NA 62 86 58 47 62
2 21 17 10 17 1 2004 0 6 1 0 4
3 43 90 88 30 1 NA 100 78 62 100 78
4 52 100 82 85 1 NA 87 83 89 100 88
5 59 76 86 64 0 NA 100 93 67 94 79
6 108 75 100 85 1 2005 100 100 100 100 97
7 118 92 76 94 0 NA 55 51 51 60 53
# i 1 more variable: emp_q26 <dbl>
샘플 R script
library(tidyverse)
# import data
helping <- read_csv("data/altruism.csv")
# rename
helping <- helping |>
rename(ph1 = pho_1, ph2 = pho_2, ph3 = pho_3)
# delete reponses
helping <- helping |>
filter(!id == 5)
# scoring
helping["phone"] <- helping |>
select(ph1:ph3) |>
rowMeans(na.rm = TRUE)
helping["persp"] <- helping |>
select(emp_q20, emp_q22, emp_q24:emp_q26) |>
rowMeans(na.rm = TRUE)
# substitute anolamies
helping <- helping |>
mutate(age = ifelse(age > 2004, 2004, ifelse(age < 2002, NA, age)))
# factors and etc.
helping <- helping |>
mutate(
sex = factor(sex, levels = c(0, 1), labels = c("male", "female")),
age = 2023 - age
)
# select variables
helping <- helping |>
select(id, sex, age, phone, persp)
정리된 파일로 분석 시작!
# A tibble: 119 x 5
id sex age phone persp
<dbl> <fct> <dbl> <dbl> <dbl>
1 1 female 19 95 75
2 2 male 20 60 58.4
3 3 NA 20 66.7 68.8
4 4 female 19 72.7 71.2
5 6 male 19 91.7 75
6 7 female 19 85.7 82.6
# i 113 more rows